
Physiology Based PK/PD Modeling.
Novel approach for drug development
PBPK / PD modeling has been extensively applied: to quantitatively translate in vitro data, predict the in vivo performance, and ultimately support waivers of in vivo clinical studies.
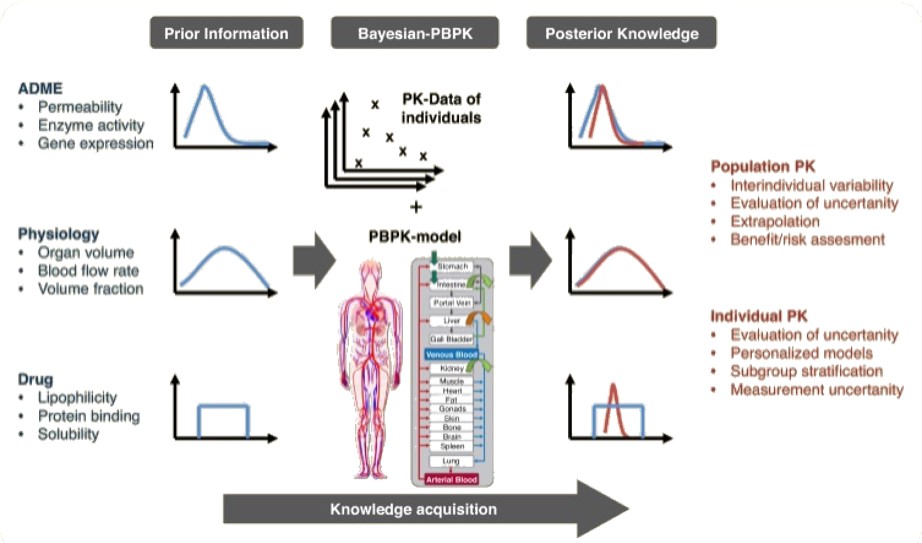
PK Models Via PBPK
Metrics Research Offers its services of OPEN-SOURCE (PK-Sim ® & MoBI ®) software to create physiologically-based QSP platforms as Insilico Modeling & Simulation solutions to support Pharmaceuticals, Drug Discovery & Development Institutes and Regulatory Authorities for decision making process along the entire life cycle of pharmaceutical products from research to the clinical use of the product.
Member & Collaborator

PBPK Capability
PBPK Consulting
We provide services for standard applications of PBPK models using the Open-Systems-Pharmacology Suite with PK-Sim ® and MoBI ®.
Services are:
- Determination of Pharmacokinetic Profile in Healthy & Diseased Virtual Human Population
- Population Variations (American, European, Asian, Black etc.)
- DDI Investigations
- First in Man Dose Predictions
- Preclinical/clinical base PBPK model development
- Special populations: PBPK-based pediatric investigations and organ impairment studies


Metrics Research have resource experiencing PBPK in Pakistan using
Below are the projects on which we have worked till now, Published!
- Enzyme and Transporter Kinetics for CPT-11 (Irinotecan) and SN-38: An Insight on Tumor Tissue Compartment Pharmacokinetics Using PBPK (view article)
- Physiologically based pharmacokinetic modeling for predicting Irinotecan exposure in human body (view article)
- Prediction of Clearance in Neonates and Infants (≤3 months of age) for Drugs which are Glucuronidated: A Comparative Study between Allometric Scaling and Physiologically Based Pharmacokinetic Modeling
(view article) - Prediction of Clearance and Dose of Midazolam in Preterm and Term Neonates: A Comparative Study Between Allometric Scaling and Physiologically Based Pharmacokinetic Modeling (view article)
